|
||
|
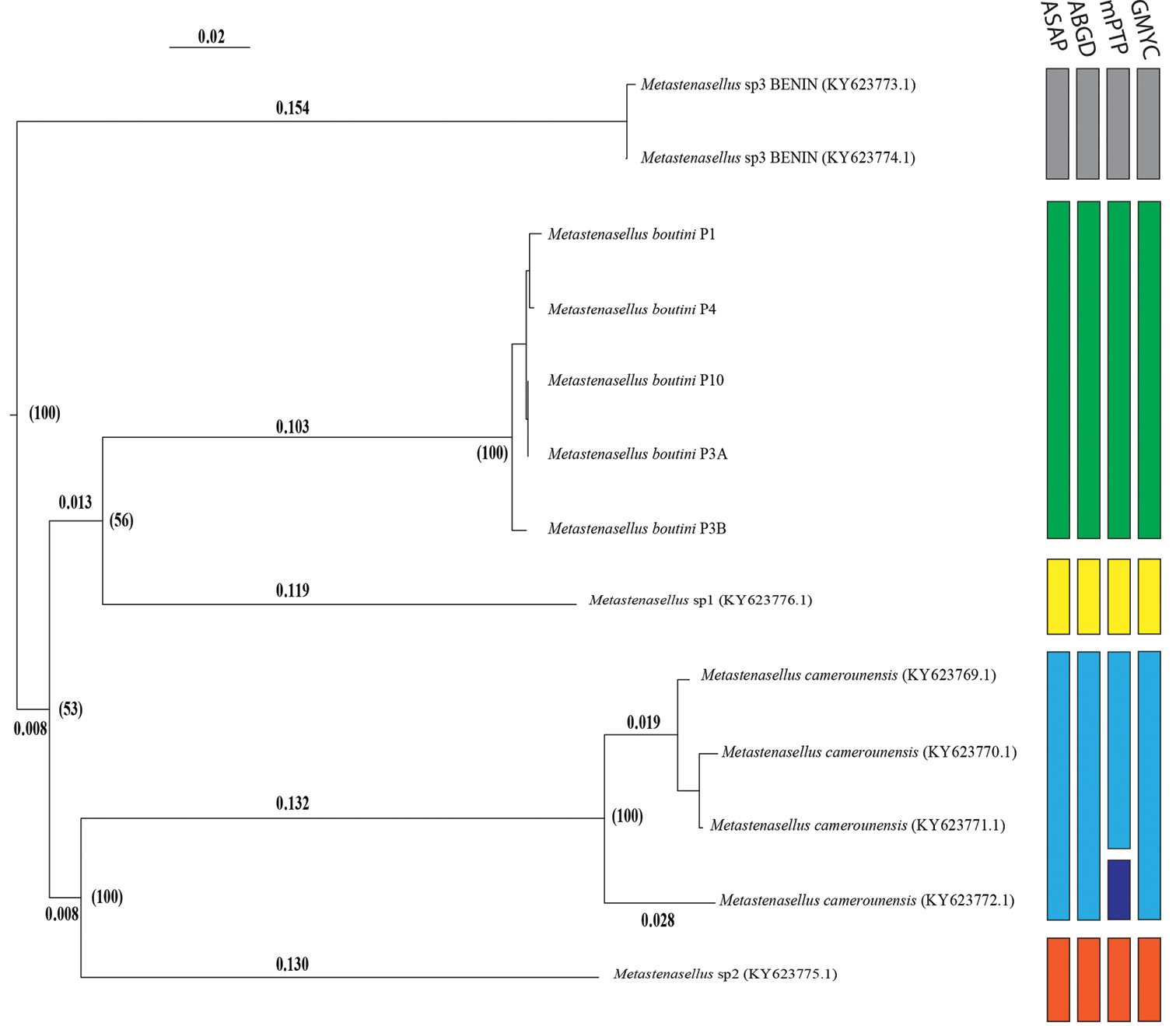
Neighbor-joining tree of the identified COI gene haplotypes. The evolutionary distances were computed using the Kimura two-parameter (K2P) model. Numbers between brackets in front of the nodes indicate bootstrap support (1,000 replicates). Boxes on the right indicate the best partition of species using ASAP, ABGD, mPTP and GMYC delimitation methods. |